
Song Wu
Ph.D. in Bioinformatics specializing in cancer alternative splicing and multi-omics data integration. Full-stack Engineer leveraging AI and computational biology to advance precision oncology and therapeutic discovery.
Education
My academic journey and formal qualifications.
Ph.D. in Bioinformatics
2019.09 - 2025.06Beijing Institute of Genomics, Chinese Academy of Sciences
China National Center for Bioinformation (CNCB)
B.Sc. in Bioinformatics
2014.09 - 2019.06Harbin Medical University
Featured Projects
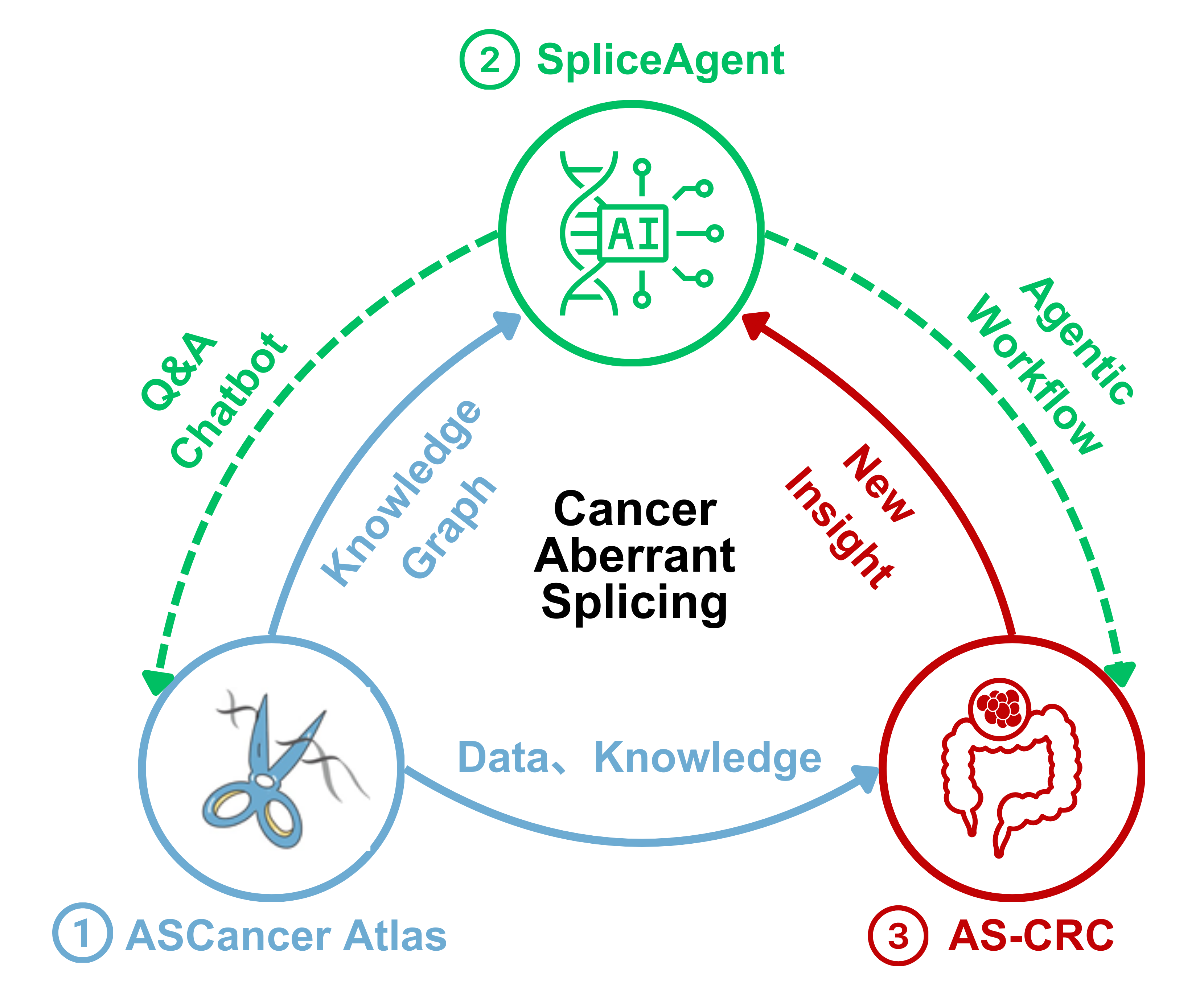
Cancer Aberrant Splicing Study
Established a comprehensive research framework (Data Resource-Intelligent Tool-Mechanism Analysis) for systematic research on cancer alternative splicing, encompassing ASCancer Atlas, SpliceAgent and AS-CRC.
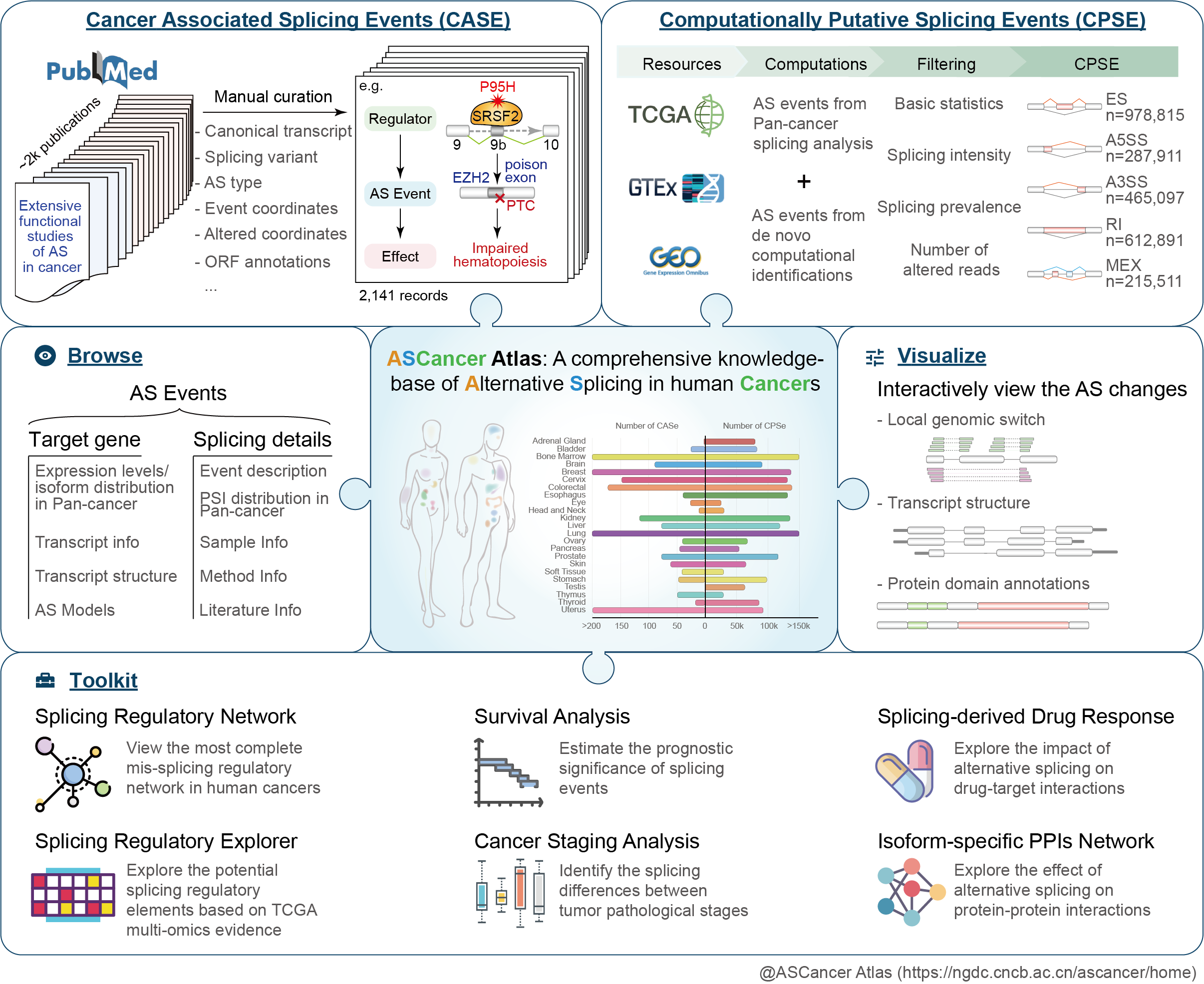
ASCancer Atlas
VisitIn this study, I compiled experimentally validated cancer-associated splicing events and systematically analyzed large-scale datasets from TCGA pan-cancer and GTEx pan-tissue cohorts to characterize splicing patterns. Additionally, I developed multi-dimensional online tools that integrate multi-omics and clinical data to identify splicing regulators, assess clinical relevance, and evaluate functional impacts through drug and protein interaction analyses. This resource equips researchers with comprehensive knowledge, data, and tools to understand cancer-specific splicing dysregulation.
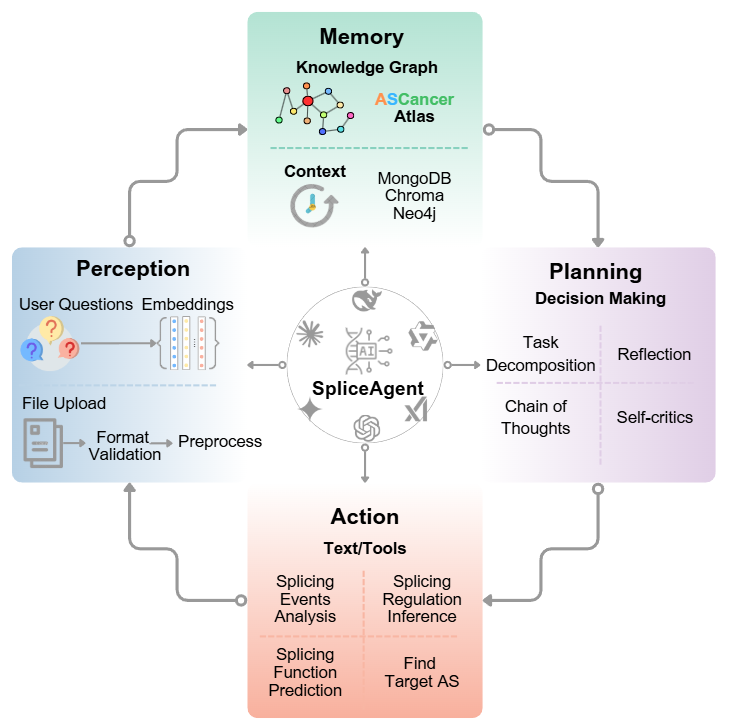
SpliceAgent (Ongoing)
VisitIn this study, I exploratively applied large language models (LLMs) and AI Agent technologies to cancer aberrant splicing research, developing the SpliceAgent. It features three core modules: 1) an intelligent curation assistant based on LLMs; 2) a cancer splicing Q&A chatbot integrating RAG, Graph-RAG, and real-time web retrieval; 3) an agentic analysis workflow automating data-to-report processes.
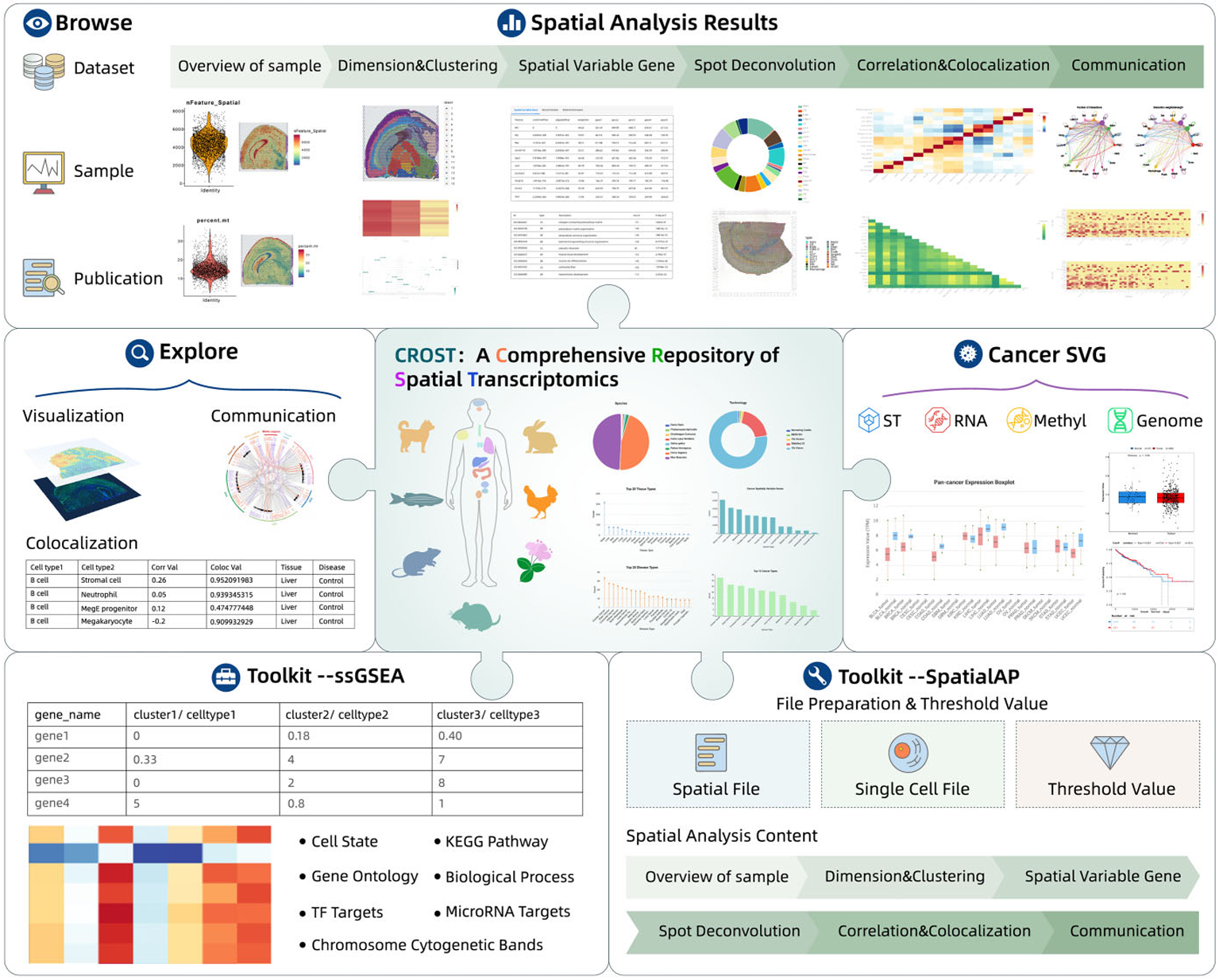
CROST
VisitIn this study, I gained extensive expertise in spatial transcriptomics through analyzing over 1,000 diverse samples across multiple spatial technologies. By creating a unified analytical platform for these datasets, I significantly enhanced research capabilities for investigating tissue structure and spatial gene expression patterns in life sciences.
First / Co-first Author (8)
(#: Co-first Author)
ASCancer Atlas: a comprehensive knowledgebase of alternative splicing in human cancers.
Wu S#, Huang Y#, Zhang M, et al.. Nucleic Acids Res, 2023, 51: D1196-D1204.
CROST: a comprehensive repository of spatial transcriptomics.
Wang G#, Wu S#, Xiong Z#, et al.. Nucleic Acids Res, 2024, 52: D882-D890.
HALL: a comprehensive database for human aging and longevity studies.
Li H#, Wu S#, Li J#, et al.. Nucleic Acids Res, 2024, 52: D909-D918.
TargetGene: a comprehensive database of cell-type-specific target genes for genetic variants.
Lin S#, Wu S#, Zhao W, et al.. Nucleic Acids Res, 2024, 52: D1072-D1081.
EryDB: A Transcriptomic Profile Database for Erythropoiesis and Erythroid-related Diseases.
Zheng G#, Wu S#, Zhang Z#, et al.. Genomics, Proteomics & Bioinformatics, 2024.
Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2023-2025.
Wu S# as co-first author in CNCB-NGDC Members and Partners.. Nucleic Acids Res, 2023-2025.